| Gene Analysis Service |
|
|
| Genotyping Analysis |
We undergo analysis by HybProbe method.
We make custom-made optimization for HybProbe design from genomic-DNA sequence
which including targeted SNP. And we will synthesize it.
After we verify that the HybProbe function is working using genomic DNA,
we will analyze SNP of your sample and report the results.
|
| Example |
|
| Analysis SNPs of Integrin α2 gene C807T (Thromb Haemost 2001;85:226-30) |
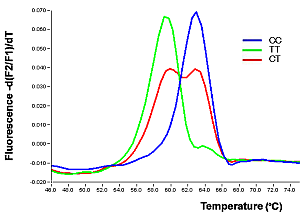 |
|
| Samples of possible contract analysis |
| Materials | Conditions of collection * 1 | Preservation Method * 2 | Way of Mailing | Send-out volume |
| Blood | Please collect blood by EDTA blood collection tube | Cold Strage
(Strage Period: within 7days) |
Cold Strage | more than 200µL |
Frozen Strage
(Strage Period: within 8days) | Frozen Strage |
| Oral Mucosa | Collect sample with swab. Please scratch with swab more than 5 times. | Frozen Strage | Frozen Strage |
- |
| hair root | Pull out hair with unpolluted hand and put it into the tube. Please confirm that the hair has hair root. |
Frozen Strage | Frozen Strage | more than 5 tubes |
| Cell | Eliminate the supernatant after collecting cells by centrifuge. | Frozen Strage | Frozen Strage |
more than 106cells |
| Tissue | - | Frozen Strage | Frozen Strage | more than 10mg |
| DNA | - | Frozen Strage | Frozen Strage | Concentration 10ng/µL
Volume more than 30µL |
| * 1 Please consult us if you collect the samples by the method other than the above. |
| * 2 Please store the samples immediately after collection. |
|
| Report Pattern |
| We will submit report and analysis data. |
 |
| |
| Quantitative Analysis of Gene Expression |
|
We determine target genes and internal standard genes ( house-keeping genes & so on).
We reverse-transcribe entrusted RNA samples and analyze them with real-time PCR. We correct level of between the samples and report more correct expression of target genes as relative values |
| - Quantitation Method of real-time PCR - |
|
When performing quantitation by real-time PCR, level of expression of the target genes is calculated out based on the Cycle Threshold (Ct Value).
When comparing level of mRNA expression of target genes between the samples, we amplify target genes by real-time PCR and obtain Ct Value, then undergo comparative quantitation.
It is a premise for performing correct quantitation. that cell number of samples are the same,
efficiency of extraction and reverse-transcription of RNA is the same and sample RNA has to be amplified by the same efficiency. However, it is difficult to satisfy all conditions actually,
so we correct the level of expression between the samples based on the quantified values of in-house standard genes( Relative Quantitation).
|
 It is possible to undergo analysis under either condition by raw samples ( tissues and cells ), total RNA and cDNA. It is possible to undergo analysis under either condition by raw samples ( tissues and cells ), total RNA and cDNA. |
 We use resultful LightCycler® and reagents of Roche Diagnostics K.K.. We use resultful LightCycler® and reagents of Roche Diagnostics K.K.. |
| |
| Application by Quantitative Analysis of Gene Expression |
We have constructed the type of analysis specific to Alternative splicing Variant besides general quantitation analysis.
As an example, following is the explanation for construction of specific detection system of VEGF121 which is the Alternative Splicing Variant of human VEGF-A.
Below scheme shows exons which constitute Alternative Splicing Variant.
|
| Result of construction of the type of detection system specific to VEGF121. |
| Exon Number |
| | 1 | 2 | 3 | 4 | 5 | 6a | 6b | 6c | 7a | 7b | 8a | 8b |
| VEGF206 | ■ | ■ | ■ | ■ | ■ | - | - | ■ | - | ■ | ■ | - |
| VEGF183 | ■ | ■ | ■ | ■ | ■ | ■ | - | - | - | ■ | ■ | - |
| VEGF165 | ■ | ■ | ■ | ■ | ■ | - | - | - | - | ■ | ■ | - |
| VEGF148 | ■ | ■ | ■ | ■ | ■ | - | - | - | ■ | - | ■ | - |
| VEGF121 | ■ | ■ | ■ | ■ | ■ | - | - | - | - | - | ■ | - |
| VEGF165b | ■ | ■ | ■ | ■ | ■ | - | - | - | - | ■ | - | ■ |
| VEGF189 | ■ | ■ | ■ | ■ | ■ | - | ■ | - | - | ■ | ■ | - |
|
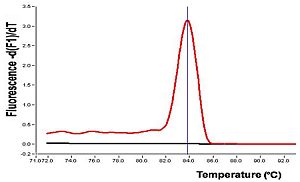 |
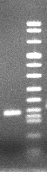
| Electrophoresis of PCR products |
| 1 M | M:Molar Weight
Marker
1:VEGF121
specific PCR |
 |
|
|
Multiple Alternative Splicing Variant are existing in human VEGF-A and multiple of them are expressed in mixed state depending on different tissues.
For this reason, it is necessary to design primers in the region specific to VEGF121 in order to detect it specifically.
Thus, we can design the system to detect VEGF121 specifically under these special conditions. |
| You can use it for the purposes other than the above. |
| · Confirmation of Efficacy of RNAi |
| · Confirmation of level of gene expression obtained from DNA microarray. |
| · Confirmation of gene expression of knock-out mouse. |
| |
| Detection Principle |
| Detection method can be chosen from Intercalater( SYBRGreen I)Method, HybProbe Method and Hydrolyzed Probe method. |
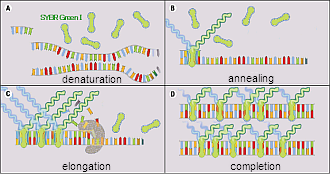 | 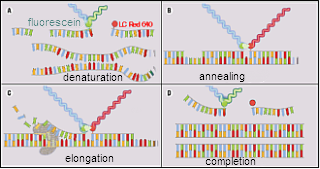 |
| Principle of Intercalater( SYBRGreen I)Method. | Principle of HybProbe Method. |
|
| |
| Conditions of Submission |
| Materials | Way of Mailing | Send-out Volume |
| Tissue | Frozen strage(DRY ICE) | about 20mg |
| Blood | Please consult us in advance. | |
| Cultured Cell | Frozen strage(DRY ICE) | more than 1 x 105cells |
| Parafine segment | Cold strage(4℃) | more than 5 pcs. of 5µm thick |
| TotalRNA | Frozen strage(DRY ICE) | more than 5µL
(concentration more than 500ng/µL) |
| cDNA | Frozen strage(DRY ICE) | more than 20µL |
|
|
| |
| Report Pattern |
|
We submit report, analysis( corrected values according to in-house standard genes).
|
 |
|
| |
| RNA Extraction · DNA Extraction · Synthesis of cDNA |
We will provide you with various Genome DNA、Total RNA、cDNA of high purity from various biological samples.
Samples are tissues, cultured cells, blood, serum, feces, paraffin segment, plant, and bacteria so on.
We are happy to undertake extraction from other samples than above.
Extracted products can be used for real-time PCR and microarray. |
| Send-out Volume |
| Sample Type | Send-out Volume |
| tissue *1 | 50mg - 100mg |
| cells *2 | 1 x 107cells |
| Total RNA *3 | more than 5µg
Concentration: more than 100ng/µL |
*1 Please freeze the tissues immediately after collection or treat with preservative reagents.
*2 Wash cells with PBS and eliminate the supernatant, then freeze it immediately in liquid nitrogen in pellet state.
*3 Please send the samples after confirming the quality by electrophoresis.
|
| Report Pattern |
| Report |
| Measured data of concentration, retio, Picture of GEL Electrophoresis |
Sample
| DNA Case: | : | TE Buffer solution more than 50µg |
| Total RNA Extraction | : | RNase free Aqueous Solution more than20µg |
| cDNA Synthesis: | : | Reverse-transcribed products of otal RNA 1µg |
* Total RNA will be delivered in frozen state and DNA and cDNA in cold storage(4℃).
* We cannot supply you with those samples in case those are to be used by our contracted service.
* Volume of delivery depends on type of samples.
Please inquire beforehand in case you wish to receive desired volume.
|
 You can see methods of sending-out samples from here. You can see methods of sending-out samples from here.
|
 |
|
| Preparation of Artificial Genes · Induction of Mutation |
| In order to obtain unique proteins, it is necessary to substitute and delete amino acids by inducing mutation into multiple sites. |
| Artificial Genes |
| No samples is required to send out. |
| Induction of Mutation |
Please send samples which will become Template.
Please inquire us for kinds of samples and needed volume.
|
| Style of Delivery |
Report, Plasmid Solution(Total Volume 5µg guaranteed), Sequence Results(Waveform, Text Sequence)
* We perform sequencing of both chains and deliver the completely matched products.
|
| |
| Precautions |
- In case we prepare entire length of sequences artificially using long chain oligonucletiodes synthesized by our company, it will become artificial gene. Possible length of sequences to prepare is more than 150bp. Please inquire us for the length of sequences for ordering.
- In case we substitute, insert and delete a part of bases based on samples as Template, it will become induction of mutation.
- We deliver prepared genes by cloning to pUC118 vector.
|
| We undergo quantity tuning of prepared plasmid. Please inquire if desired. |
 |
| |
| Preparation of Expression Vector |
We insert target genes into desired expression vector and prepare the product. We supply inserted genes after confirming the base sequences.
Please consult us as we can prepare artificial genes even if it’s difficult to get Template.
|
| |
| Samples to be sent to us. |
 Template for preparation of genes (1 positive kind). Template for preparation of genes (1 positive kind). |
cDNA: volume of one synthesis of cDNA type synthesis which is commonly used(one tube of more than 20µL of liquid concentrate))
DNA:30µL by 10ng/µL
Plasmid:30µL by 1ng/µL
|
 Information on mailing samples (including sequence information) Information on mailing samples (including sequence information) |
| Please fill in the "Information on mailing samples" which will be sent at the time of receiving application and send it to us. |
| Style of Delivery |
|
* We perform sequencing of both strands and deliver the completely matched products. |
| |
| Precautions |
- Please send plasmid samples of which sequences are confirmed without fail.
Please consult us in case of other samples.
- We undergo quantity tuning of prepared plasmid. Please inquire if desired.
|
 You can see methods of sending-out from here. You can see methods of sending-out from here. |
 |
| |
| Sequence Analysis |
We will provide you with rapid and accurate Sequence Analysis Service.
|
| Basic α Course |
We will decide sequences with DNA Template and Primers sent to us.
Guaranty of Decoding 500 base sequences. |
|
Please select from the followings regarding primers.
|
- Send out primers in different container from Template.
- We will synthesize primers.
|
| |
| Advance Course |
We decide long base sequences by primer Walking Method.
Please select single-stranded or double-stranded analysis. |
| Report Pattern |
Base Sequences(Text File), Waveform File
* We attach result of analysis with mail8 ( Condensed to ZIP format ) |
| Precautions |
- We recommend more than OPC purification grade for primers to be used for sequence analysis.
( If unpurified grade, good analysis results may not be obtained)
- If purification of samples is not enough, good analysis results may not be obtained due to contamination of salt, proteins and impurities, so please purify samples good enough.
- There is a case that good results cannot be gained due to primer sequences(low Tm value, dimes are formed so on).
|
| |
 You can see methods of sending-out samples and sample volume from here. You can see methods of sending-out samples and sample volume from here. |
 Browsing method of Waveform File is from here. Browsing method of Waveform File is from here. |
 |
|
| HPV Gene Test |
| About Cervical Cancer |
| Cervical cancer is increasing rapidly in 20's-30's. |
- Cervical cancer has high occurrence frequency next to breast cancer in gynecology.
- Uterus cancer has cervical cancer develops at uterine cervix and uterine corpus cancer develops at uterus mucosa of uterus corpus.
- There is almost no subjective symptom in cervical cancer, so it is frequently delayed to find it.
- Occurrence frequency of cervical cancer is increasing year by year and 8,500 Japanese women develop it yearly in gynecology field,
and it is reported that about 2,400 die every year.
|
| (National Cancer Center , Centers for Cancer Control and Information Services) |
| Cervical cancer is mainly caused by infection of high-risk type human papilloma virus(HPV). |
- High-risk type HPV is a temporary infection in many cases and it will be eliminated naturally in many cases,
- however it may cause cervical cancer if infection lasts for a long period.
- Not only a special people get infected to HPV but it' a very common virus that a lot of women get infected during course of life.
- There are about 15 kinds of high-risk type HPV, and HPV 16 and 18 are the most frequently found type among them in cervical cancer.
|
| (Onuki M et al.:Cancer Sci100(7):1312-1316, 2009) |
